Example of hyperspectral image analysis using the software
HIA2.1
Unzip Data.zip
Data.txt: hyperspectral CARS intensity images of a U2OS cell
acquired with the multimodal CARS microscope at School of
Biosciences (Cardiff University) using spectral focusing. Software:
MultiCARS.
Offset.txt: CARS intensity image of the same cell taken at zero
temporal overlap of the Pump and Stokes pulses. Software: MultiCARS.
gwratio.dat: Glass/Water CARS ratio spectrum
1. Import hyperspectral data
- File>Load>MultiCARS
- for tiff or ascii import, check
type (CARS, SRS, Raman) and scaling (pixel size, spectral axis,
intensity scale)
2. Remove offset
- Image>Subtract Offset>Image
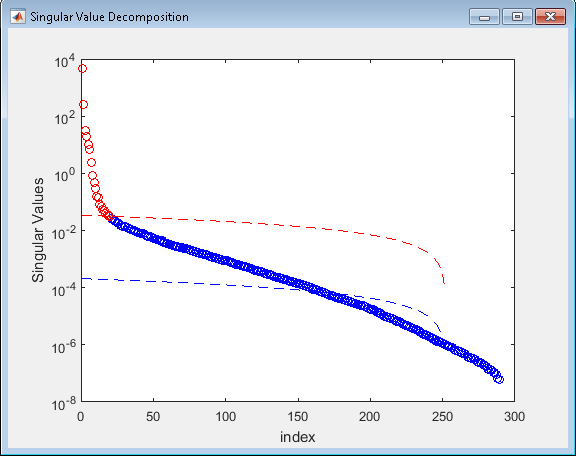
3. SVD denoising
- Analysis>SVD denoising
- SVD settings:
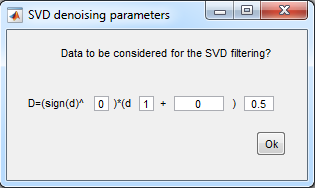
- if negative values are
present in the data, use (sign(d)^1)*(d^2+0)^0.25
- Press Ok
- Denoising settings:
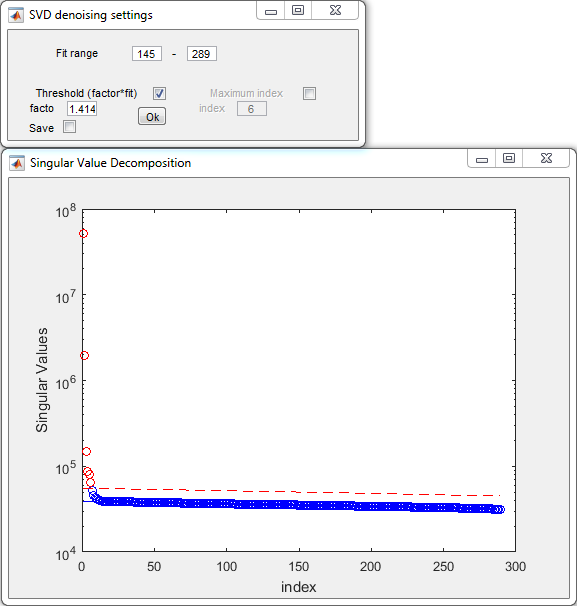
- Press Ok
- Check SVD denoised data (Data 1: original data, Data 2:
denoised data)
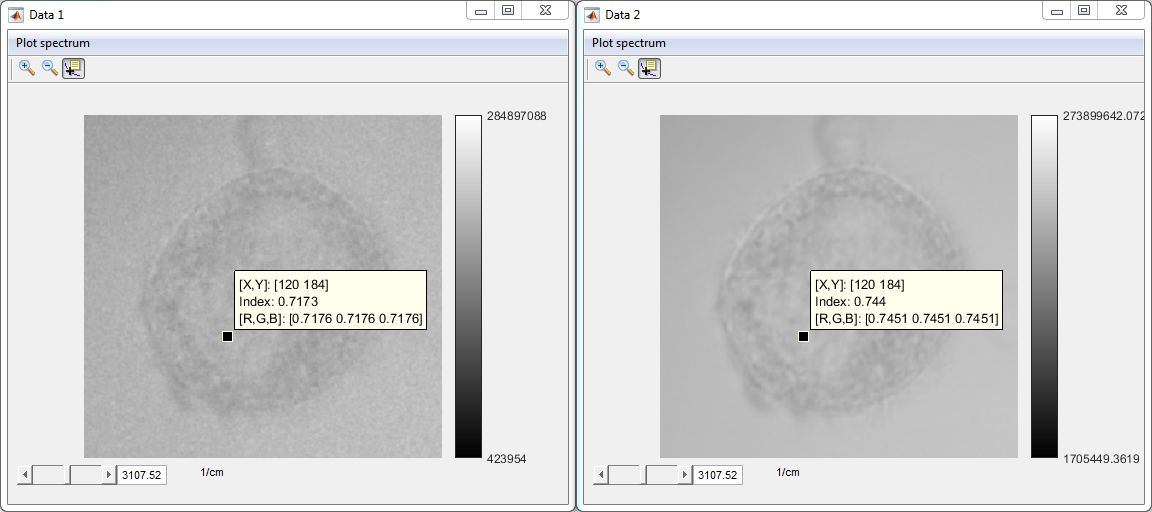
4. Calculate CARS ratio
- Analysis>CARS ratio calculation
- CARS ratio settings:
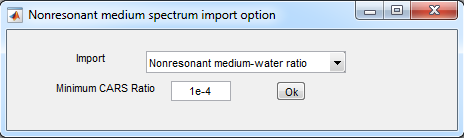
- Press Ok
- Load gwratio.dat
- Select region corresponding to water
- Type>Mask
- Selection>Freehand (double click on the selected area to
activate)
5. Correction for uneven background
- Analysis>Uneven background correction>Background region
determination>Manual
- Settings:
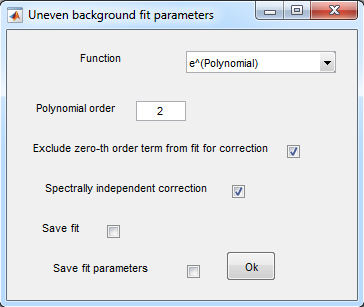
- Press Ok
- Select region corresponding to water
- Type>Mask
- Energy>Range
- Selection>Freehand (double click on the selected area to
activate) - select area similar to previous step
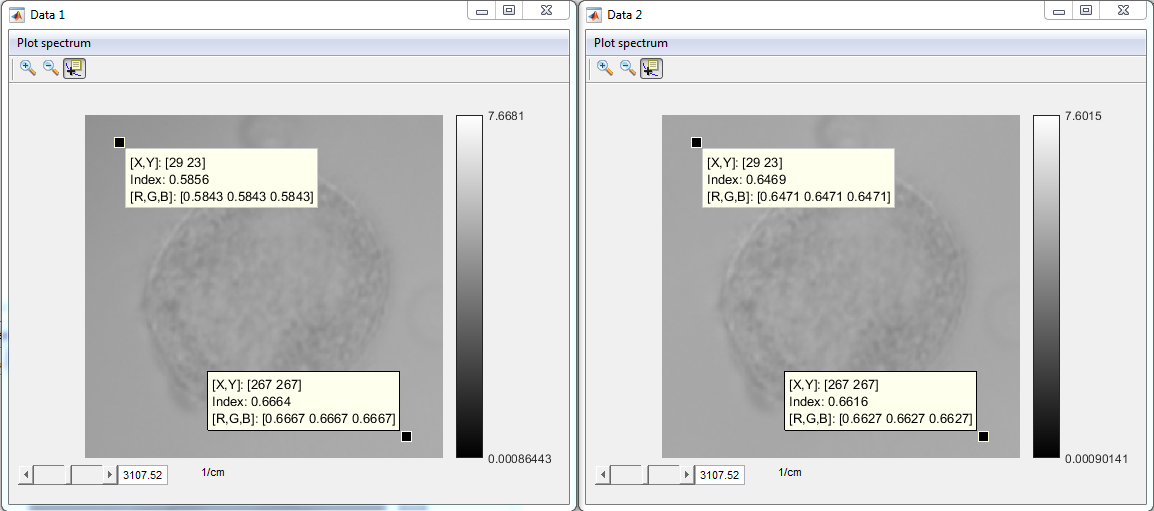
- Press Ok
- Check the results (Data 1: uncorrected data, Data 2: corrected
data)
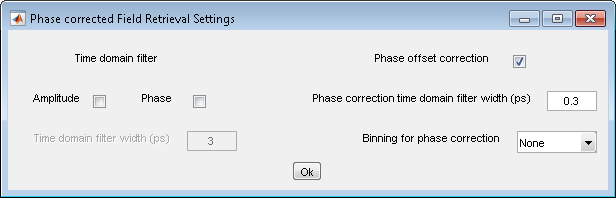
6. Field retrieval
- Analysis>Field retrieval (PCKK)
- Settings:
7. Save compressed data
- It is recommended to save the data at different step of the
analysis.
- File>Save>HIA file
- Data type: 16bit
- SVD denoising parameters:
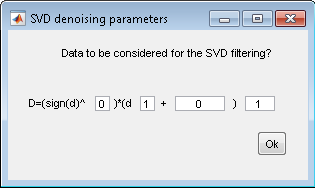
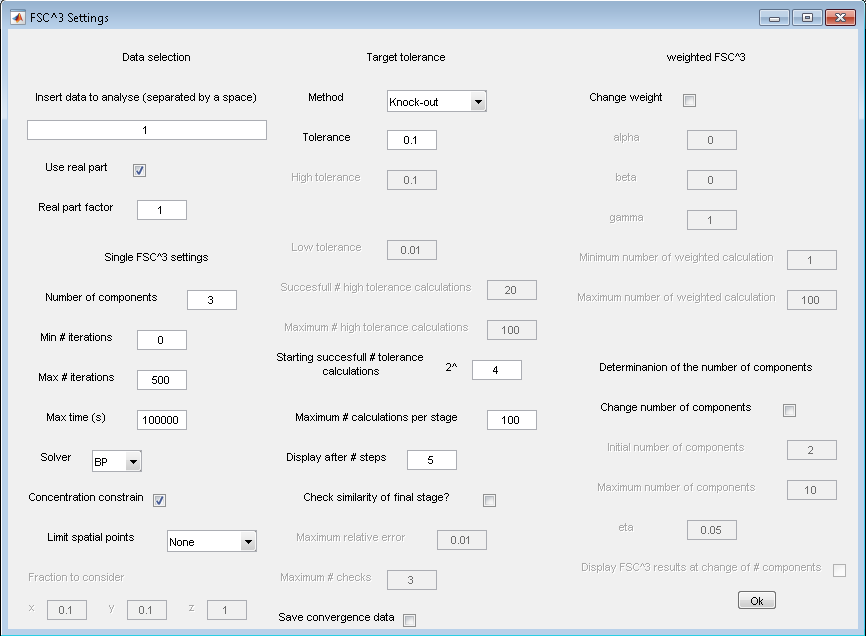
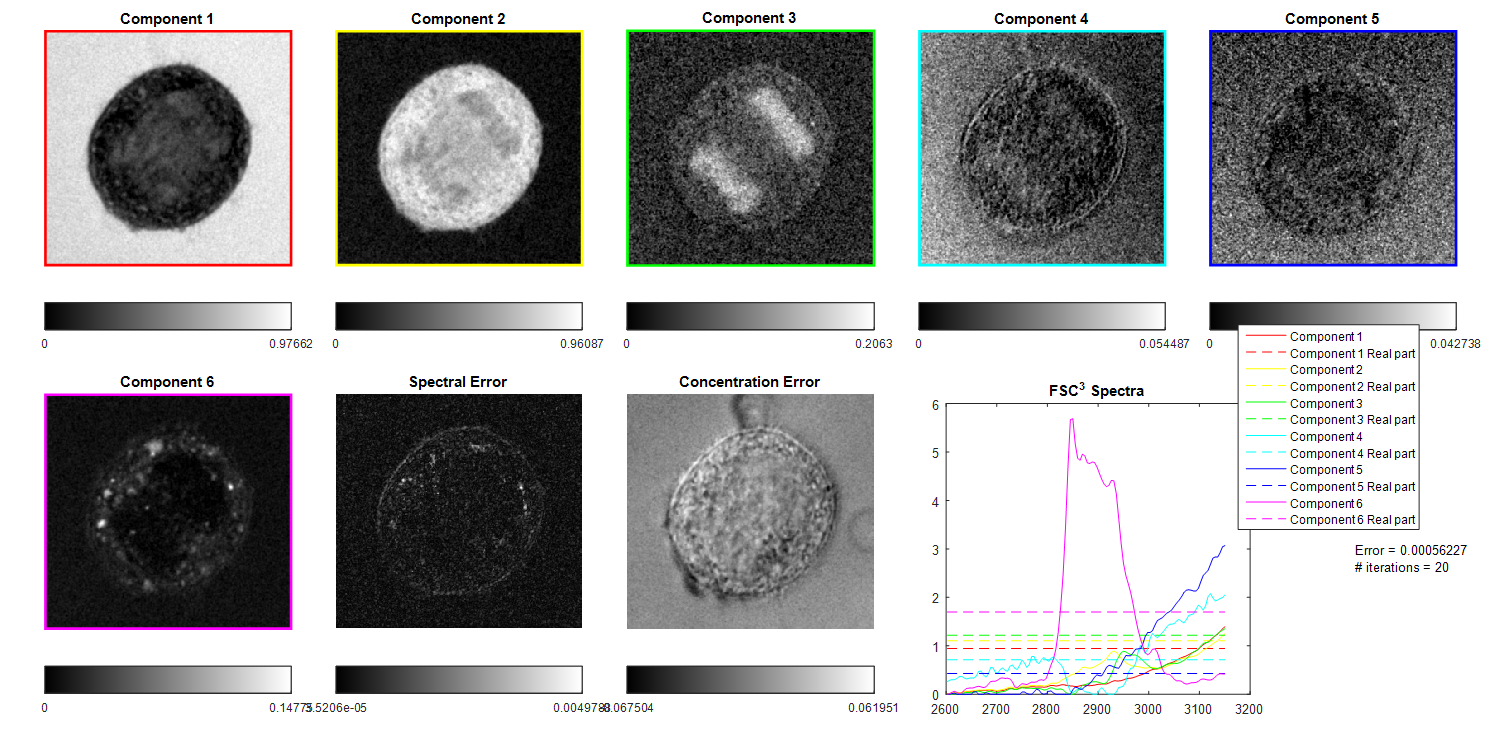
8. FSC3
- Limit spectral range (if needed)
- Image>Limit spectral range
- Limit from 2700 to 3100 1/cm
- Analysis>FSC3
- FSC3 settings
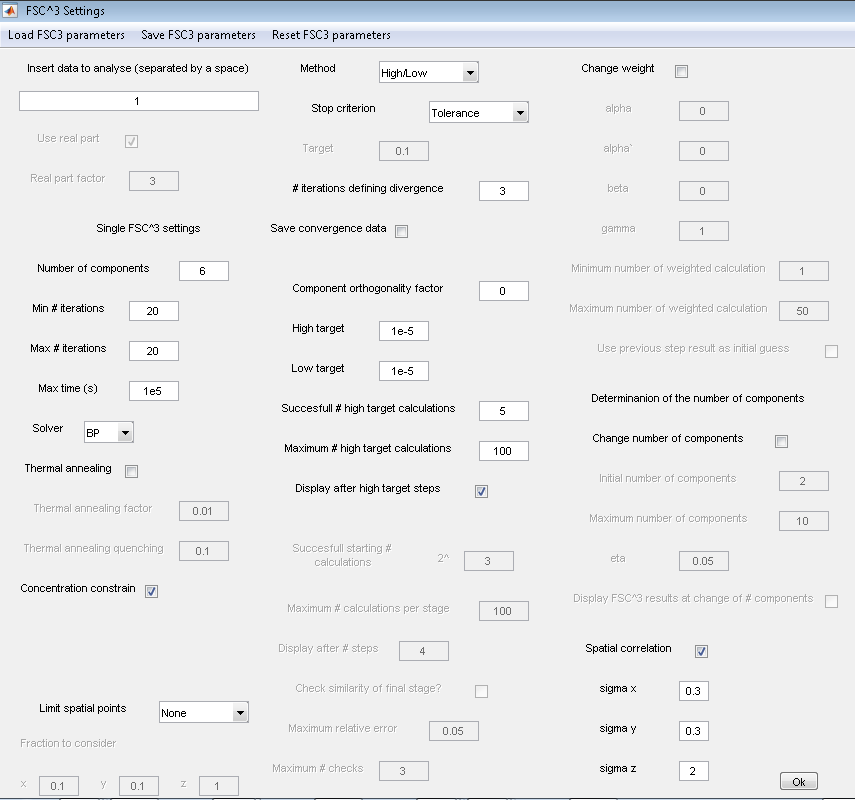
Recommended settings for improved reproducibility
We noticed improved reproducibility of the factorization of the
example data using the FSC3 settings shown below. Settings can be
imported in the v2.1 using the 'Load FSC3 parameters' function on
the FSC3 panel menu and selecting the file 'FSC3_2p1.par'.
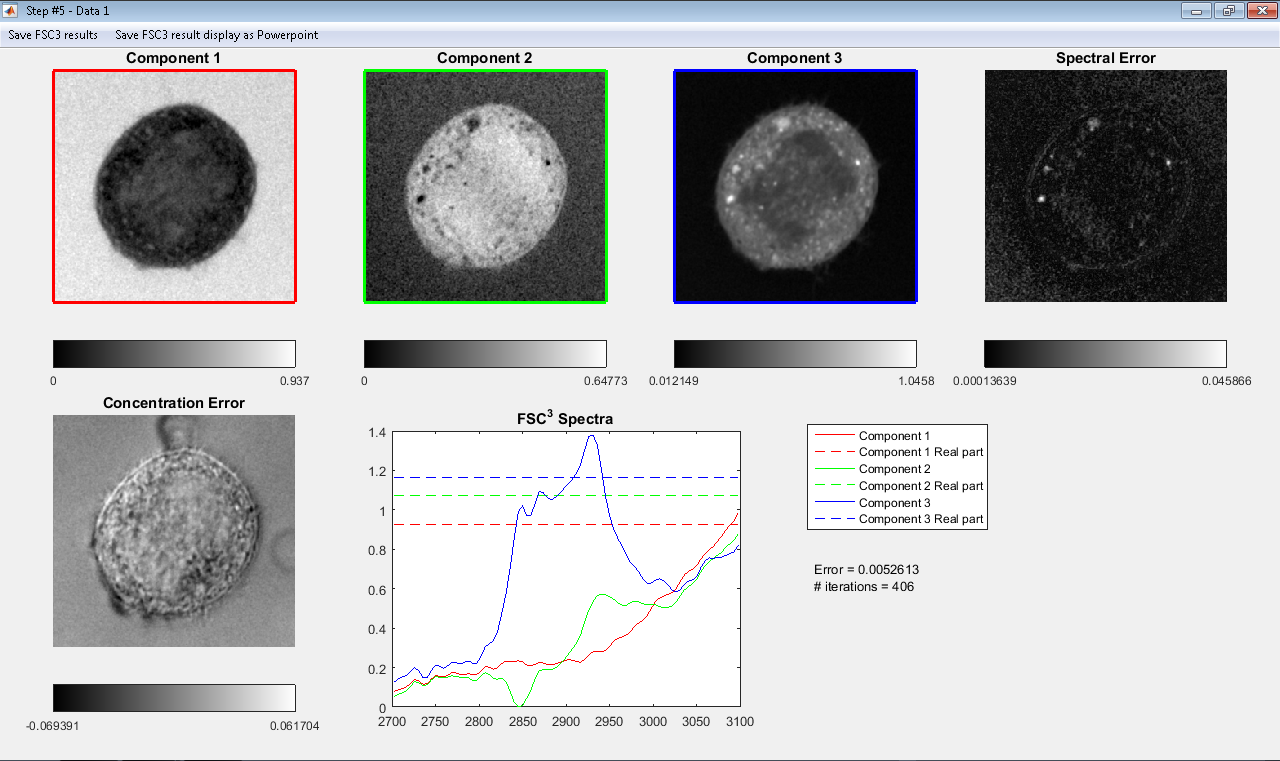
These settings produce some results were the water is factorized in
only one component and DNA and lipid are factorized separately from
the cytosolic protein.
07/05/2015, Francesco Masia. Updated by FM
26/06/2015
















